 Anglerfish
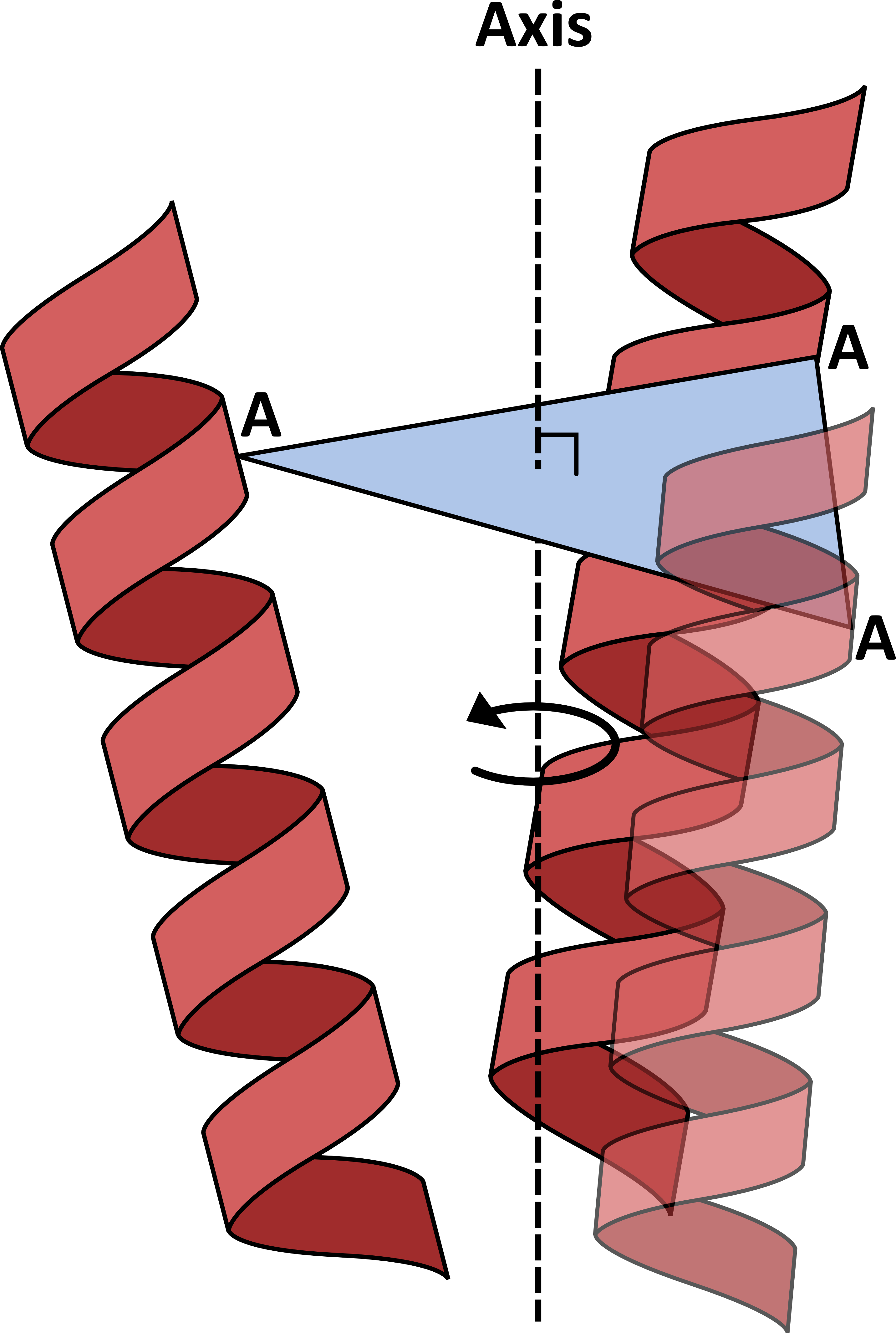
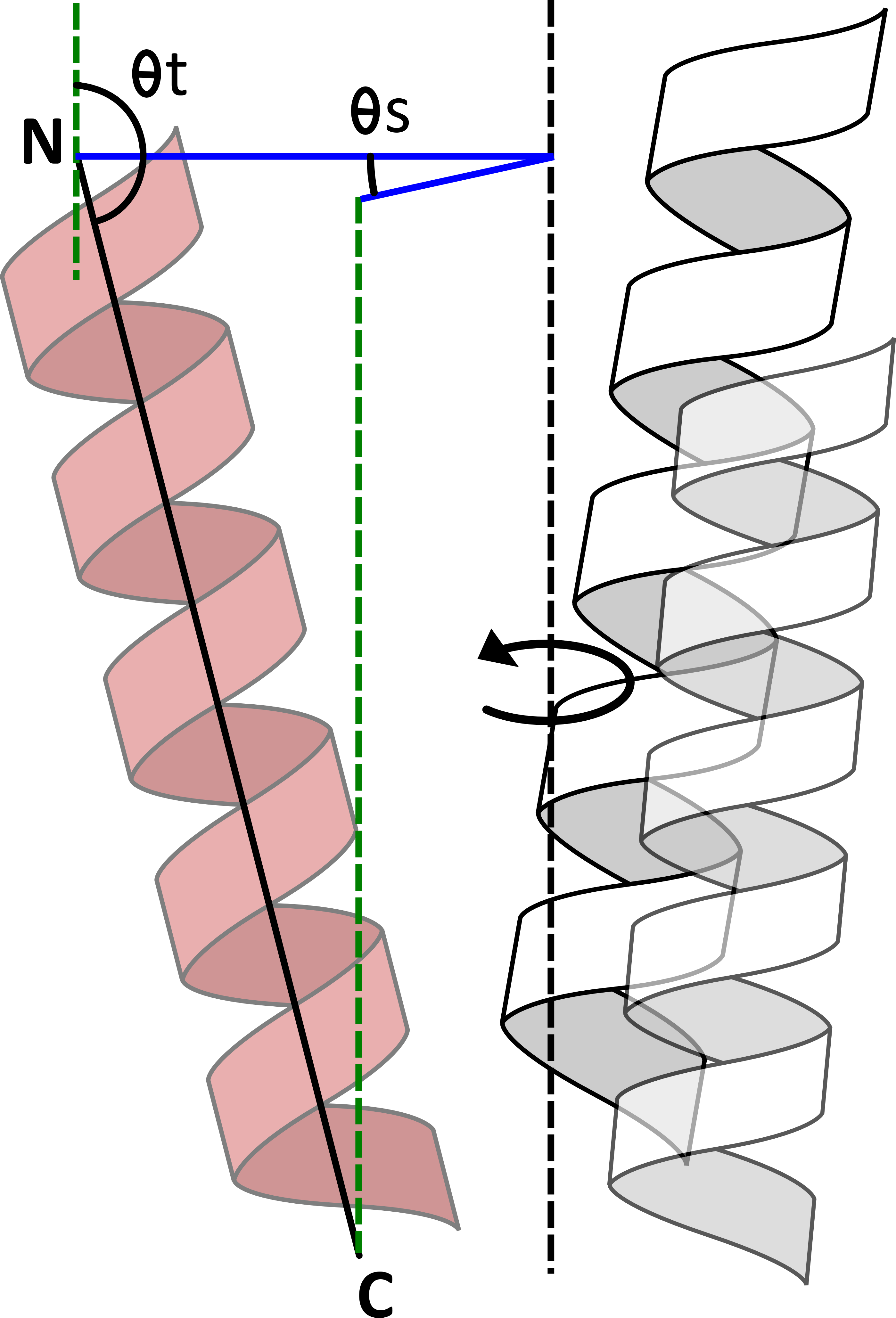
AnglerfishAnglerfish is a method of determining the orientation of an α-helix relative to a defined axis in a rotationally symmetric protein structure such as a membrane protein channel. This axis (Axis in the figure) is the axis of rotational symmetry within the structure, which is defined as the normal to a plane containing the C atoms of a given residue from each monomer (A). For example, given a channel this will be the pore axis. The helix is defined between a start residue (N) and end residue (C). The orientation is split into 2 components:
The tilt (θt): the angle between the helix and a parallel to the axis at N.
The swing (θs): the angle between the axis and C at N in the plane perpendicular to the axis.
For more information on running anglerfish see the help page or the paper: Colledge and Wallace (2016) Bioinformatics 33(8), 1233-1234
To run Anglerfish you will need: